A Binary Contrastive Learning Encoder
A Binary Contrastive Learning Encoder inspired by Google’s Supervised Contrastive Learning model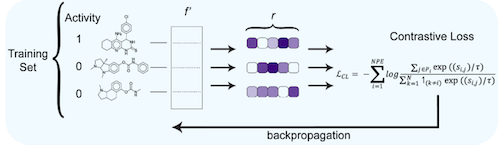
A Binary Contrastive Learning Encoder inspired by Google’s Supervised Contrastive Learning model
A solubility classification model web application
Created and containerized a SQL database using Docker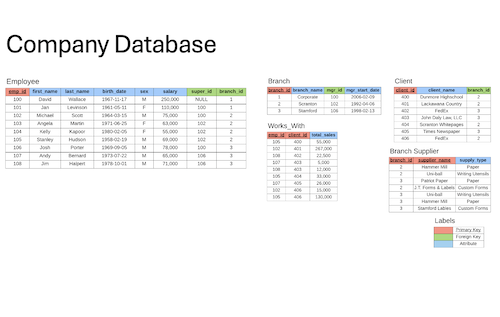
Healthcare Dashboard using Tableau
Demonstrating PySpark command to work with a dataframe and using PySpark ML to build a simple regression model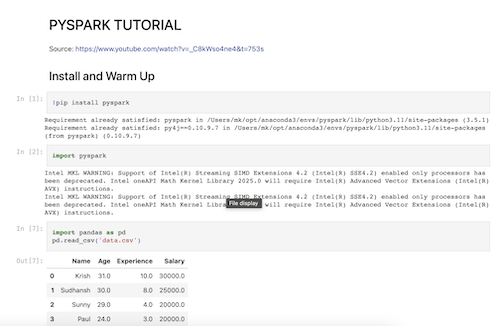
Solutions of the HackerRank SQL Problems
Published in Biomimetics, 2020
Paper on plankton dispersal within complex flow environments
Recommended citation: Ozalp, M.K. (2020). "Experiments and agent-based models of zooplankton movement within complex flow environments." Biomimetics. 5(1). http://kemalozalp.github.io/files/dispersal.pdf
Published in Journal of Chemical Information and Modeling, 2024
Paper on using sequential contrastive and deep learning models to identify selective BChE inhibitors for the therapeutic treatment of Alzheimer’s Disease
Recommended citation: Ozalp, M.K., Vignaux, P.A., Puhl, A.C., Lane, T.R., Urbina, F. and Ekins, S., 2024. Sequential Contrastive and Deep Learning Models to Identify Selective Butyrylcholinesterase Inhibitors. Journal of Chemical Information and Modeling. https://pubs.acs.org/doi/abs/10.1021/acs.jcim.4c00397
Published in , 2024
Paper on the metachronal paddling of saltwater zooplankton, brine shrimp, using computational fluid dynamics
Recommended citation: Senter, M. D. et. al. (2024). "The hydrodynamics of metachronal paddling in brine shrimp." TBD. 1(3).
Published in , 2024
Paper on the movement behavior of saltwater plankton, Artemia inferred from the statistical analysis of 3D trajetories
Recommended citation: Ozalp, M. K. et. al. (2024). "Statistical analysis of *Artemia* trajectories suggest that superdiffusive movement starts earlier than the feeding." TBD. 1(3).
Published in Nature Communications, 2024
Paper on comparing the performance of classical ML, few-shot learning and transformer-based LLM (MolBART) at varying dataset sizes
Recommended citation: Snyder, S. et. al. (2024). "The Goldilocks Paradigm: Comparing Classical Machine Learning, Large Language Models, and Few-Shot Learning for Drug Discovery Applications." Nature Communications. 1(3).
Published:
I gave this poster presentation at the Society of Integrative and Comparative Biology (SICB) conference held in Tampa, FL in January, 2019. Please check out the paper published in 2020 to learn more about my research.
Published:
I gave this presentation at the Society of Integrative and Comparative Biology (SICB) conference held in Austin, TX in January, 2020. By the time I gave this talk, my paper on the same topic had been already published. So, please check out the paper to learn more about my research.
Lab, The University of North Carolina at Chapel Hill, Department of Biology, 2017
BIOL 101L is an introductory Biology lab designed for freshman undergraduate students who study Biology. It is an examination of the fundamental concepts in biology with emphasis on scientific inquiry. Biological systems will be analyzed through experimentation, dissection, and observation.
Lab, The University of North Carolina at Chapel Hill, Department of Biology, 2018
BIOL 278L is a lab designed for sophomore and senior undergraduates study in Biology. It teaches techniques of observation and experiments in animal behavior. Students design or conduct experiments in guidance of their teaching assistants to study the behavior of various invertebrates and vertebrates throughout the course. Students also learn and perform statistical analysis methods, such as Student’s T-test, regression, and ANOVA, on the data they collected.
Undergraduate Course, The University of North Carolina at Chapel Hill, Department of Biology, 2019
BIOL 692H is a course designed senior and senior biology majors aiming for Honors and Highest Honors. In this course, students learn the preparation of a written and oral presentation of honors thesis research.